Testing the clock model in BactDating
Xavier Didelot
2025-08-07
Source:vignettes/exampleRelaxed.Rmd
exampleRelaxed.RmdInitialisation
library(BactDating)
library(ape)
set.seed(0)Data generated from strict clock model
We start by generating a coalescent tree with 10 leaves sampled at regular intervals between 1990 and 2010, and a coalescent time unit of 5 years:
dates=1990:2010
phy=simcoaltree(dates,alpha=5)
plot(phy,show.tip.label = F)
axisPhylo(backward = F)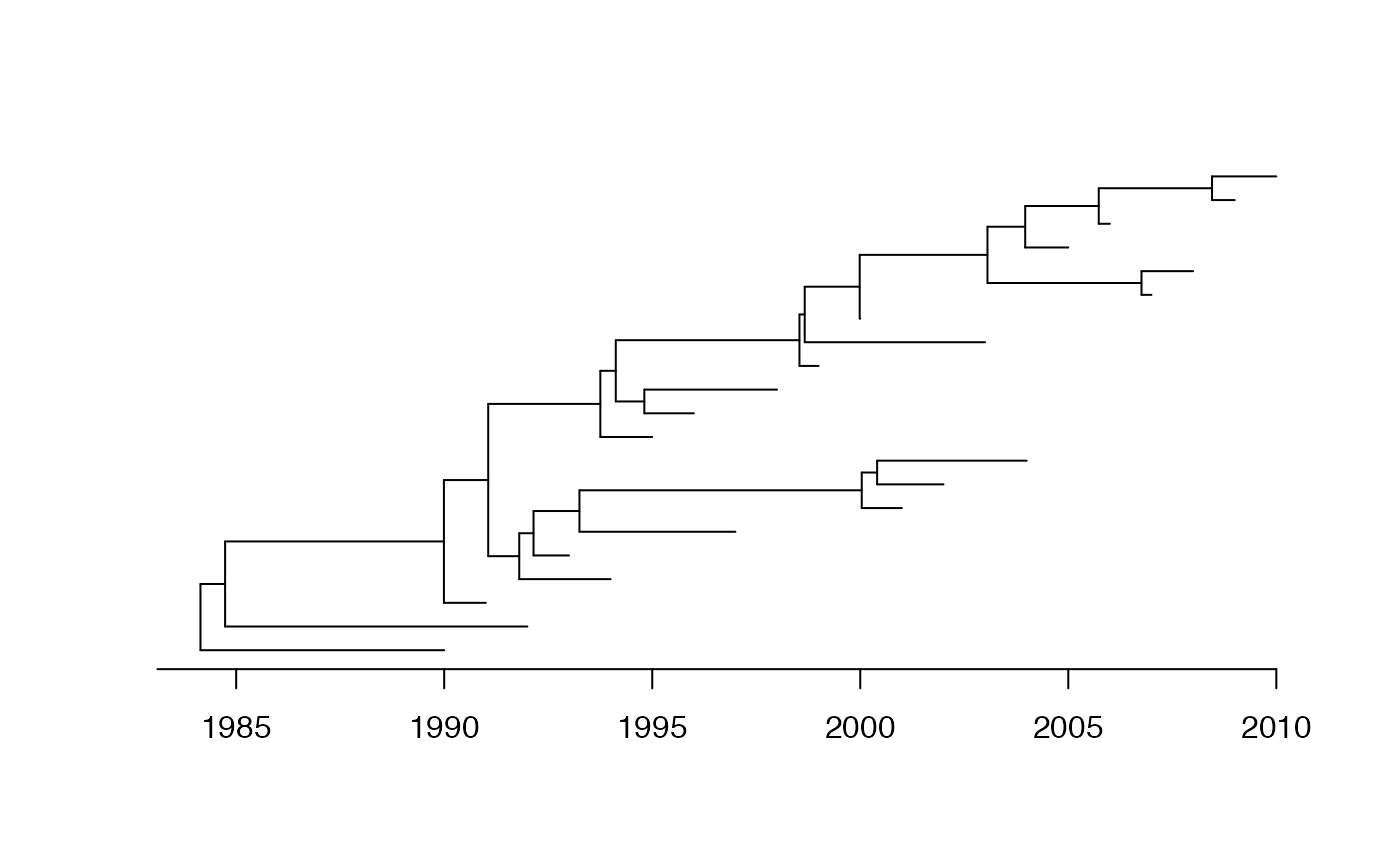
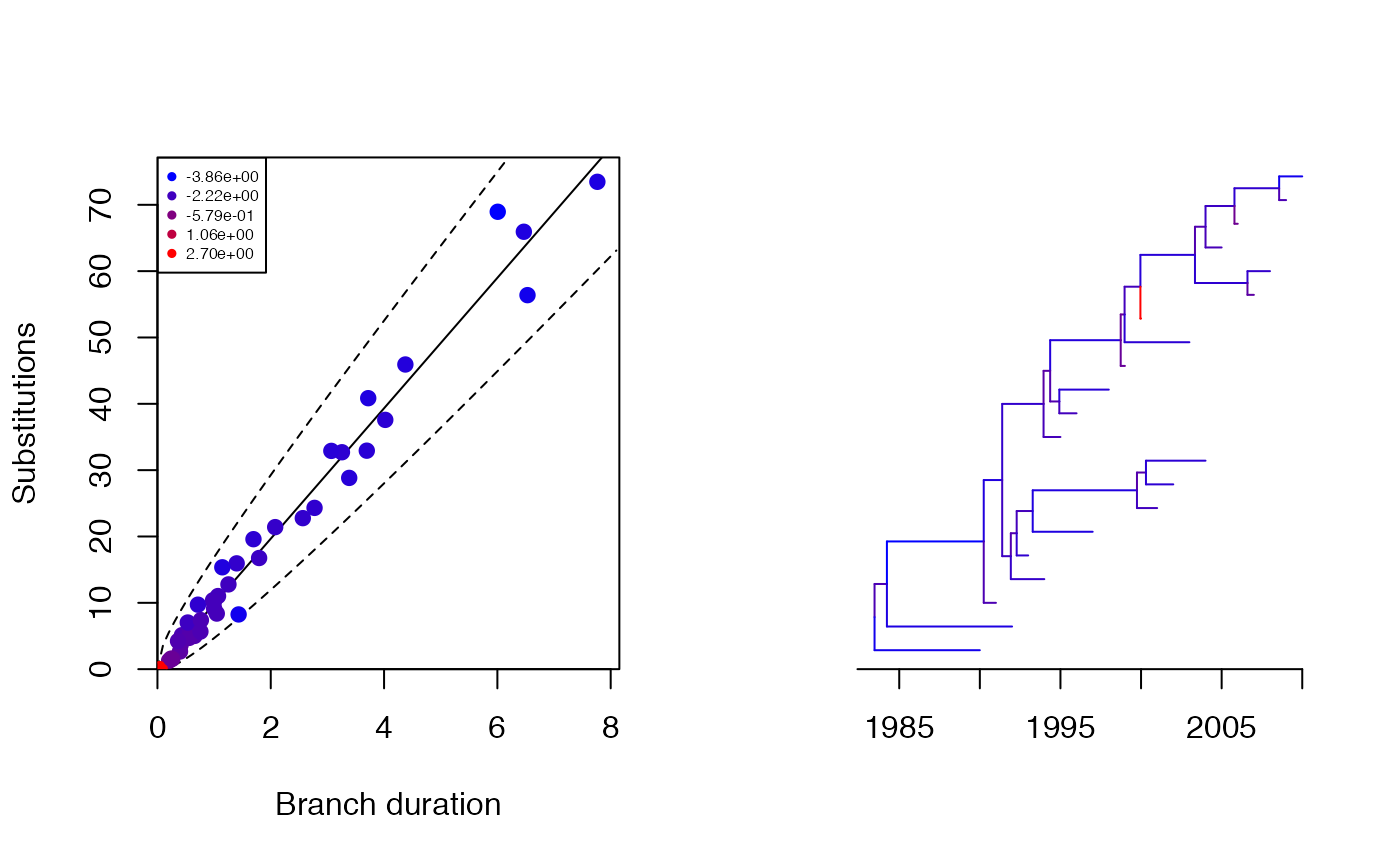
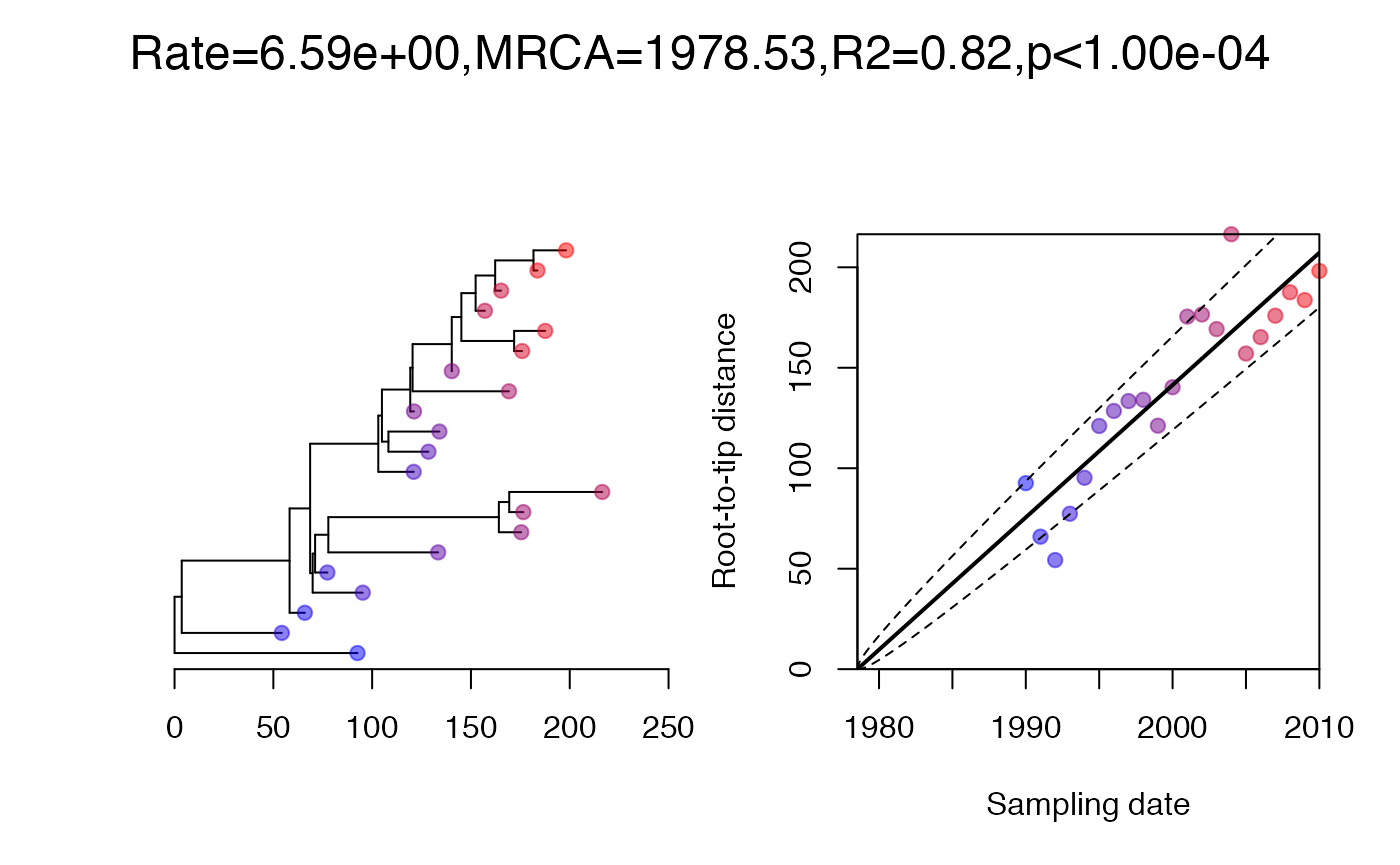
On each branch we observe a number of substitutions which is distributed where is the branch length and per year is the substitution rate. We can simulate an observed phylogenetic tree and perform a root-to-tip analysis as follows:
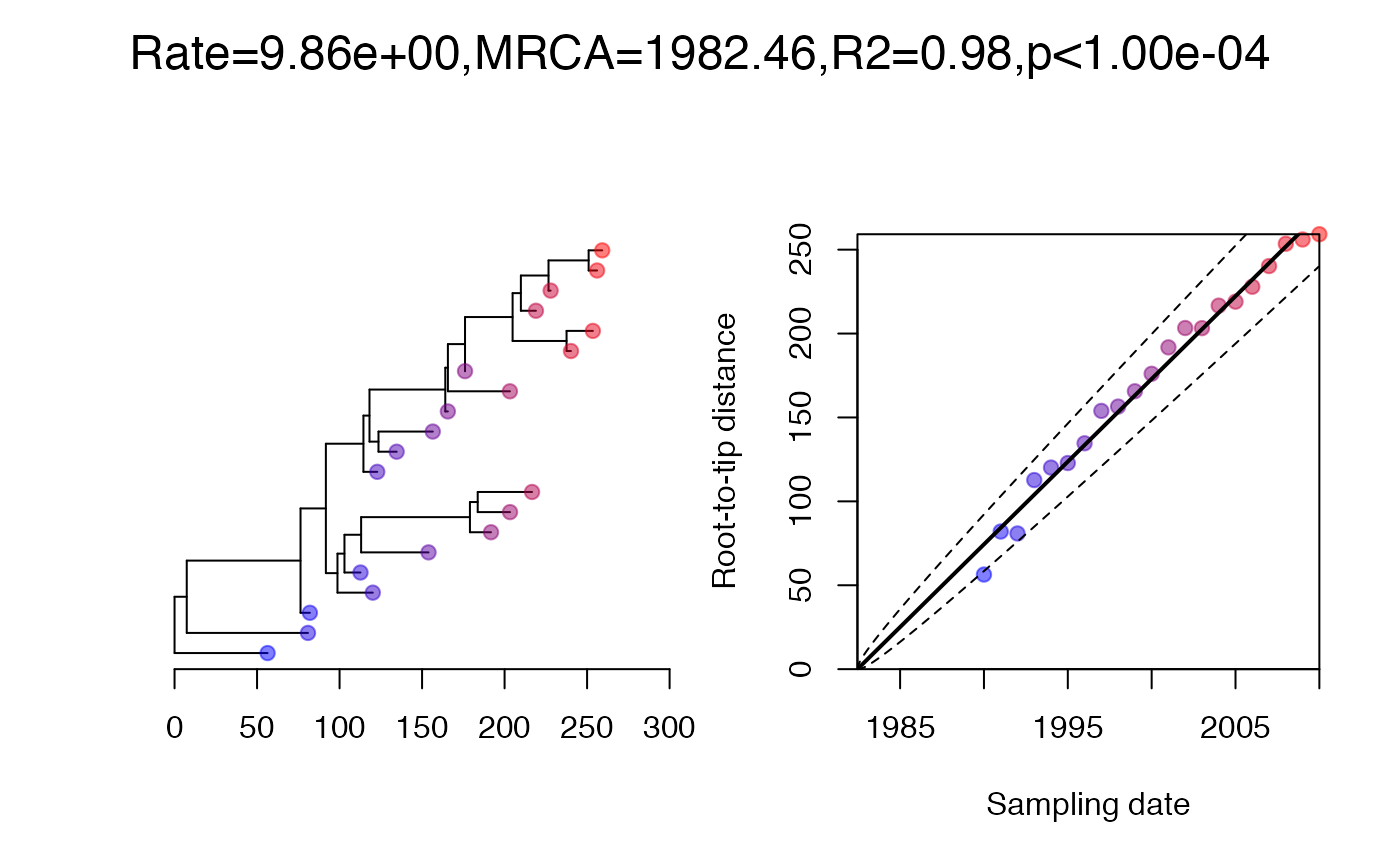
Analysis of data from strict clock model
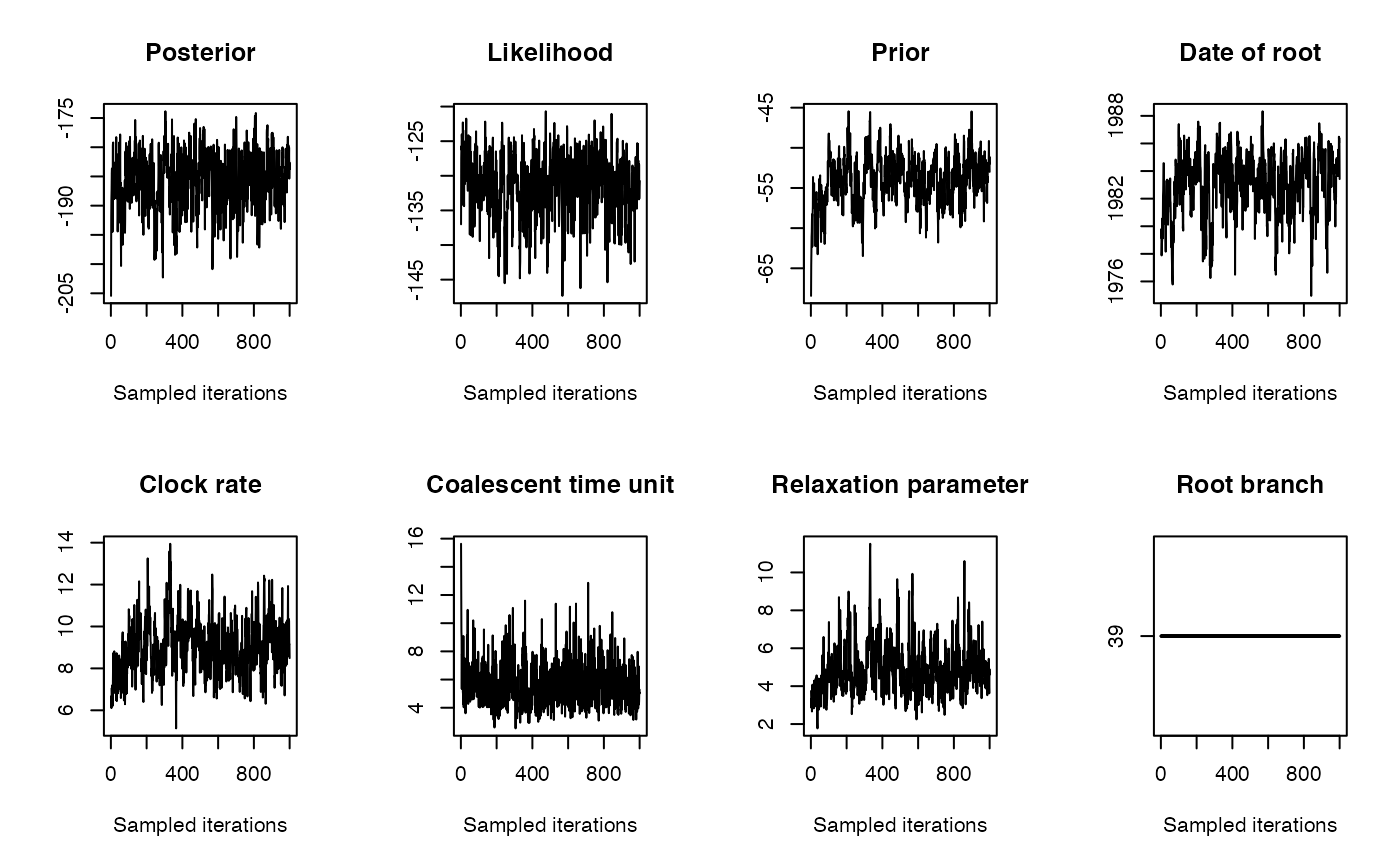
We run the dating analysis as follows:
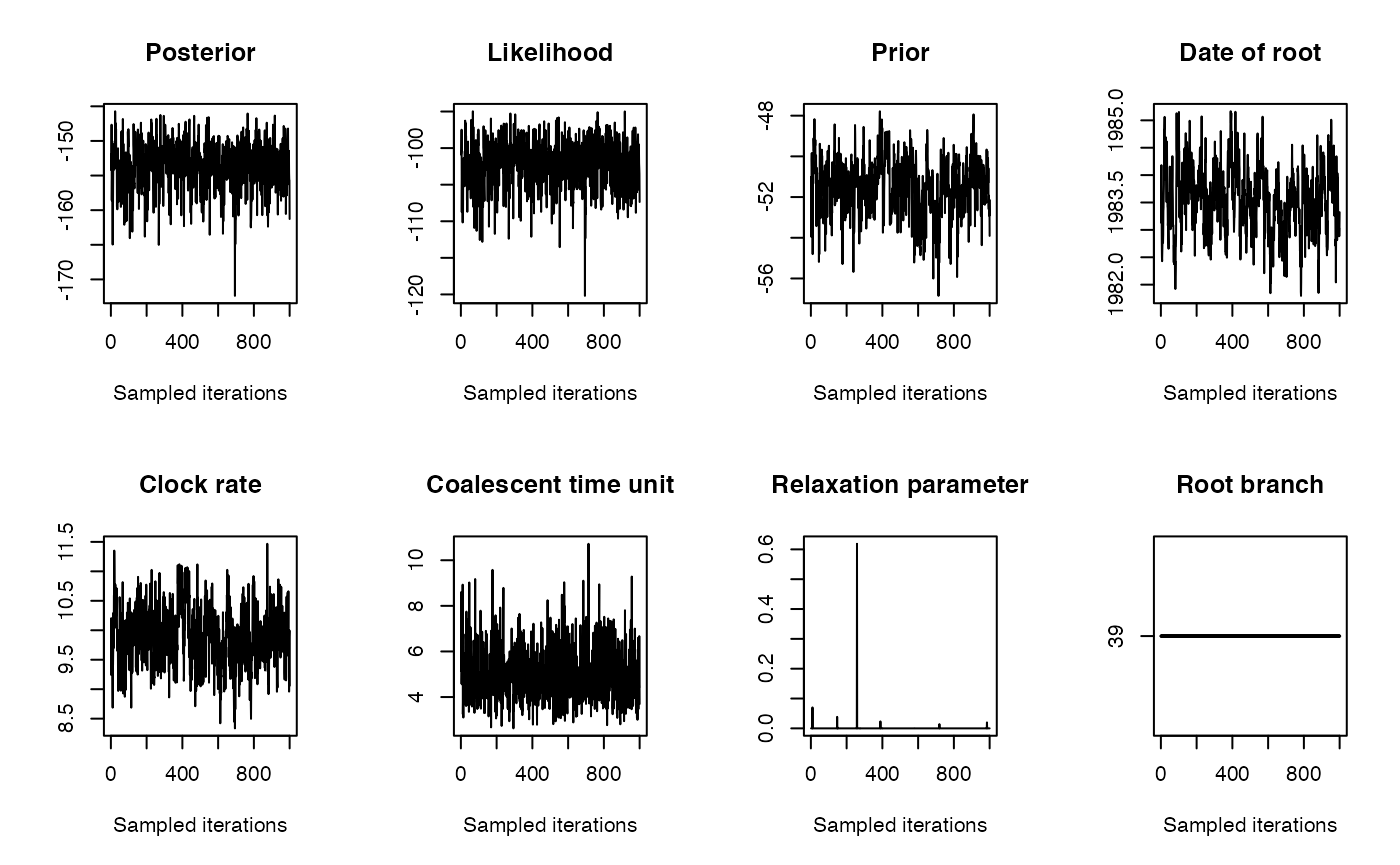
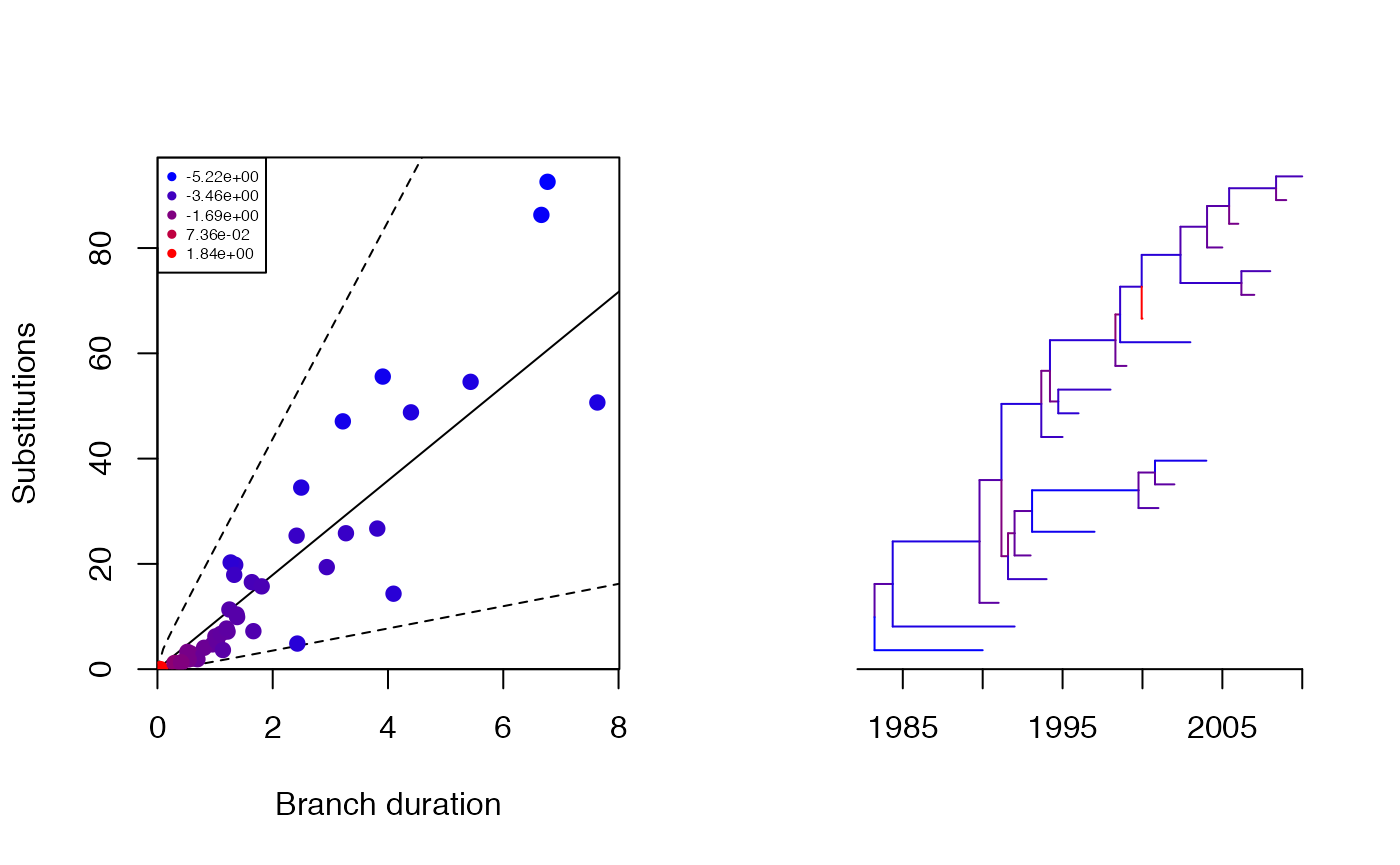
print(res$pstrict)## [1] 0.993Let’s see how each branch contributes to the overall likelihood:
plot(res,'scatter')