The clustered permutation test in BactDating
Xavier Didelot
2025-08-07
Source:vignettes/clustered.Rmd
clustered.RmdBackground
In this vignette we demonstrate the usage of the experimental
clustered permutation test implemented in the command
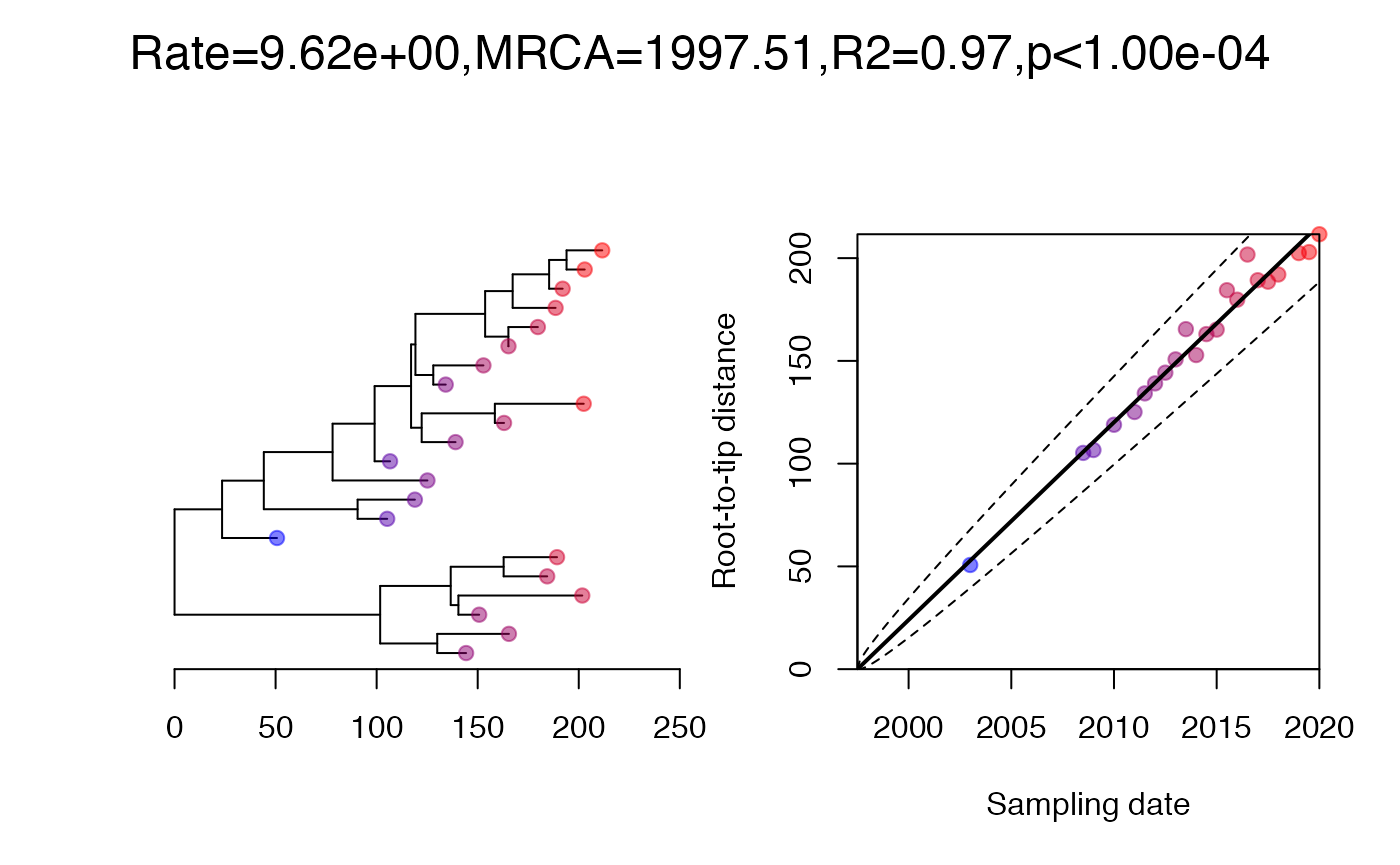
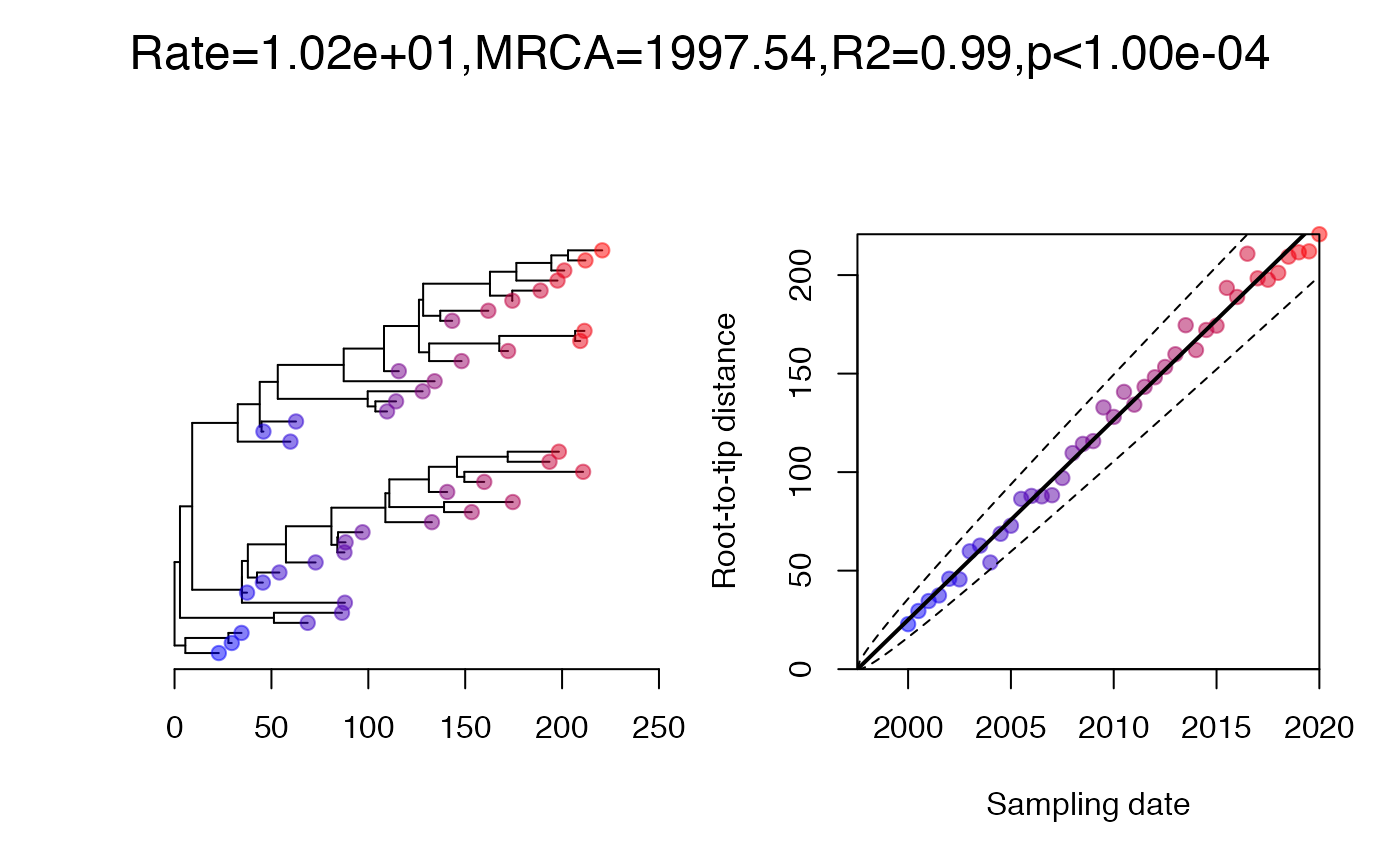
clusteredTest. Permutation tests are often used to measure
the strength of a temporal signal, and in BactDating this
is implemented in the command roottotip to calculate the
p-value of the temporal signal. However, this test can be confounded by
the presence of genetic structure and/or non-uniform sampling over time
or lineages. To account for this effect, Duchene et al (2015) proposed a
clustered permutation test, which was later refined by Murray et al
(2016). In this test, a Mantel test between pairwise temporal and
genetic distances is being used to assess the confounding effect. If
this Mantel test is initially significant, but becomes insignificant
after merging monophyletic clusters with similar dates (eg within a
year), then this clustering is enough to eliminate the confounding
signal. A clustered permutation test is then used to assess the strength
of the temporal signal, in which dates are permuted between clusters
only (ie each cluster always has the same date in the permutations).
The command clusteredTest is based on some of these same
ideas, but does not exactly replicate the procedure described above.
More details to follow.
Initialisation
library(BactDating)
library(ape)
set.seed(0)Data
We start by generating a simulated dataset:
dates=seq(2000,2020,0.5)
phy=simcoaltree(dates,alpha=10)
tree=simobsphy(phy,mu=10)
plot(phy,show.tip.label = F)
axisPhylo(backward = F)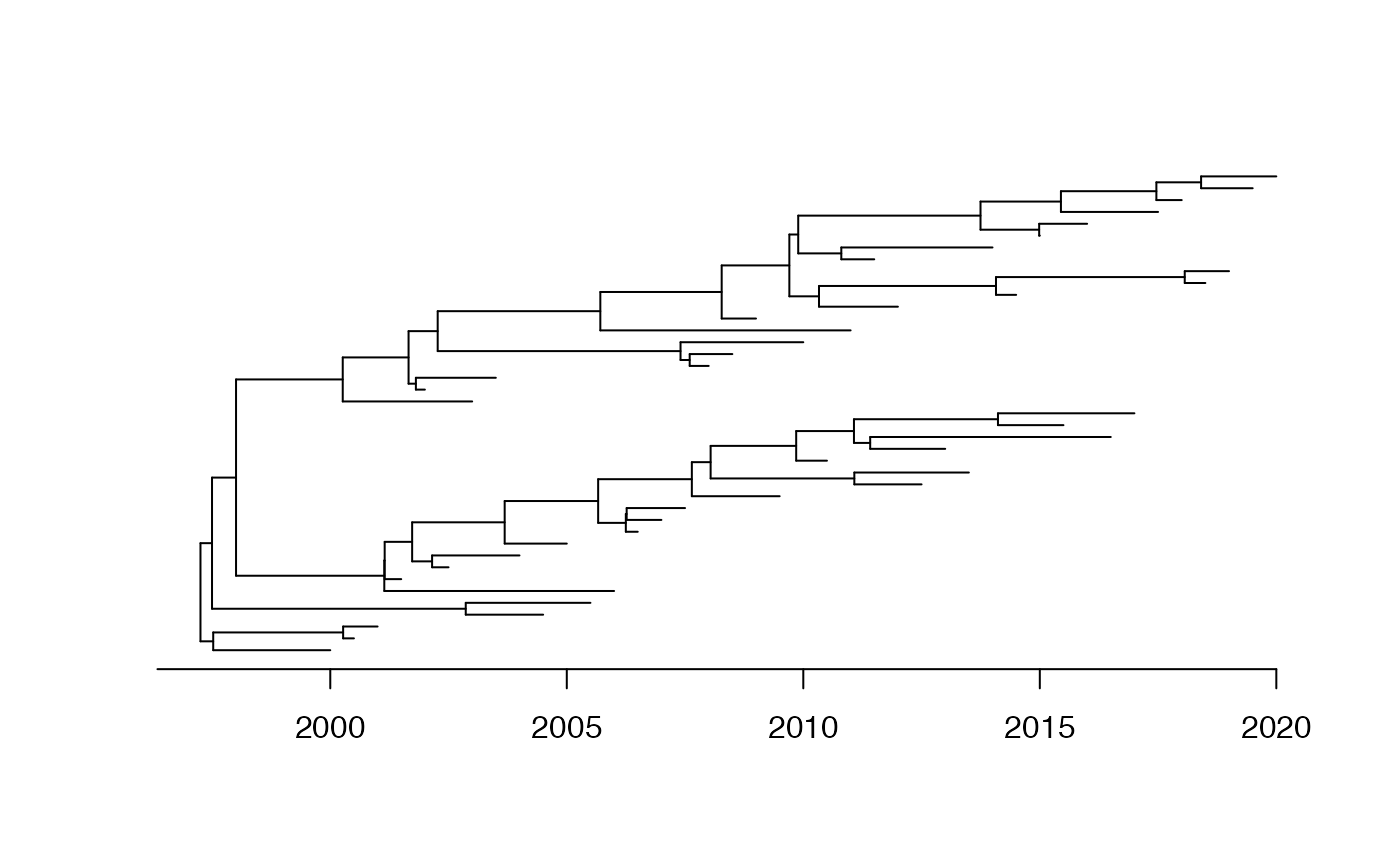
Clustered permutation test
Let’s try the clustered permutation test analysis:
res=clusteredTest(tree,dates)