Demonstration of DetectImports principle
Xavier Didelot
2025-11-19
Source:vignettes/principle.Rmd
principle.RmdSimulation with constant population size
For initial simplicity, consider that the local population has constant effective population size .
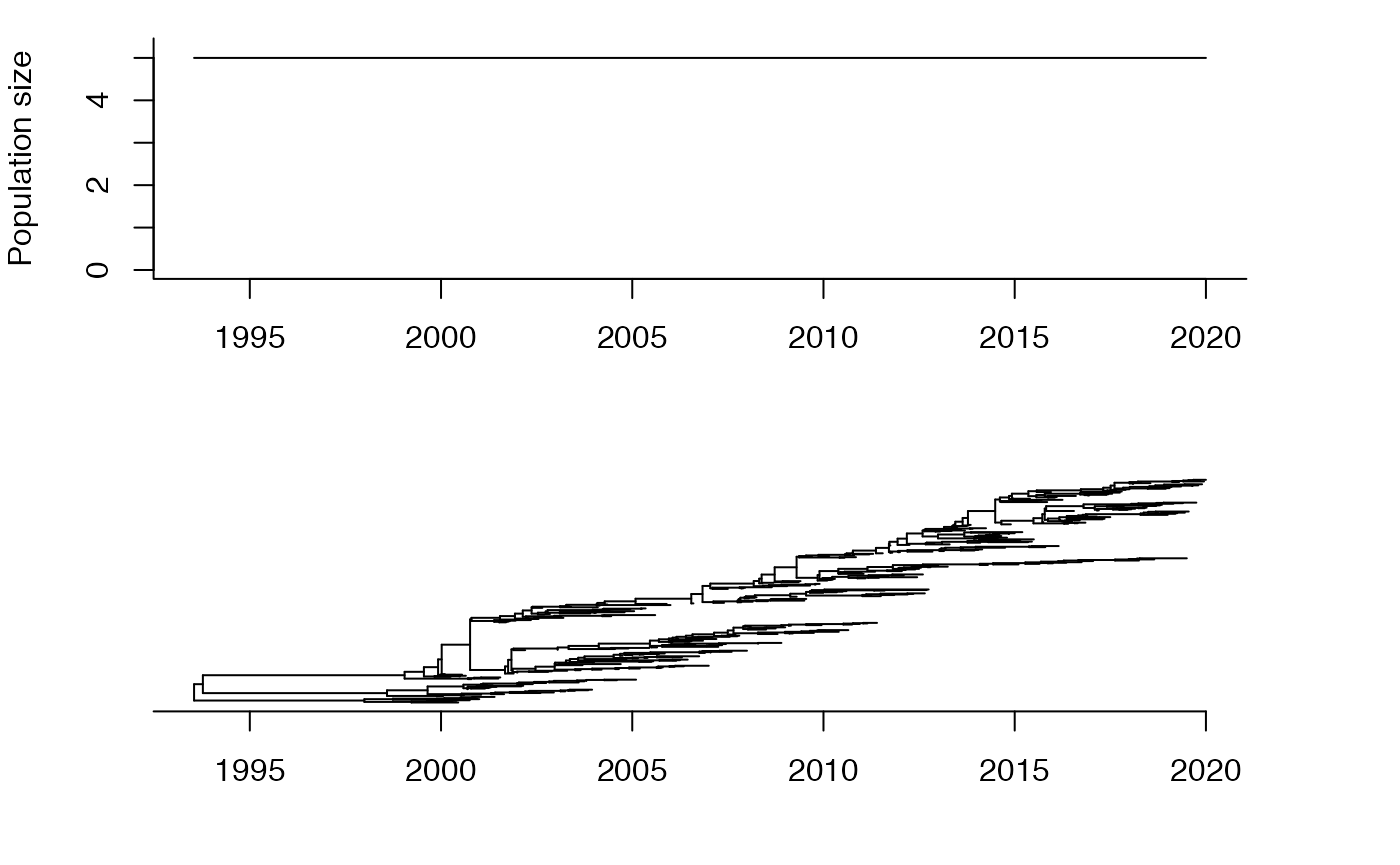
Coalescent intervals
We consider the leaves in increasing order of date. Let denote the date of leaf , we have . The -th leaf has to coalesce with the genealogy formed by the first leaves. Let denote the sum of branch lengths in the genealogy formed so far between the time of leaf and the time where it coalesces with an ancestor of a previously considered leaf. We call the ``coalescent interval’’. Let’s plot on the y-axis against on the x-axis:
plotCoalInt(tree)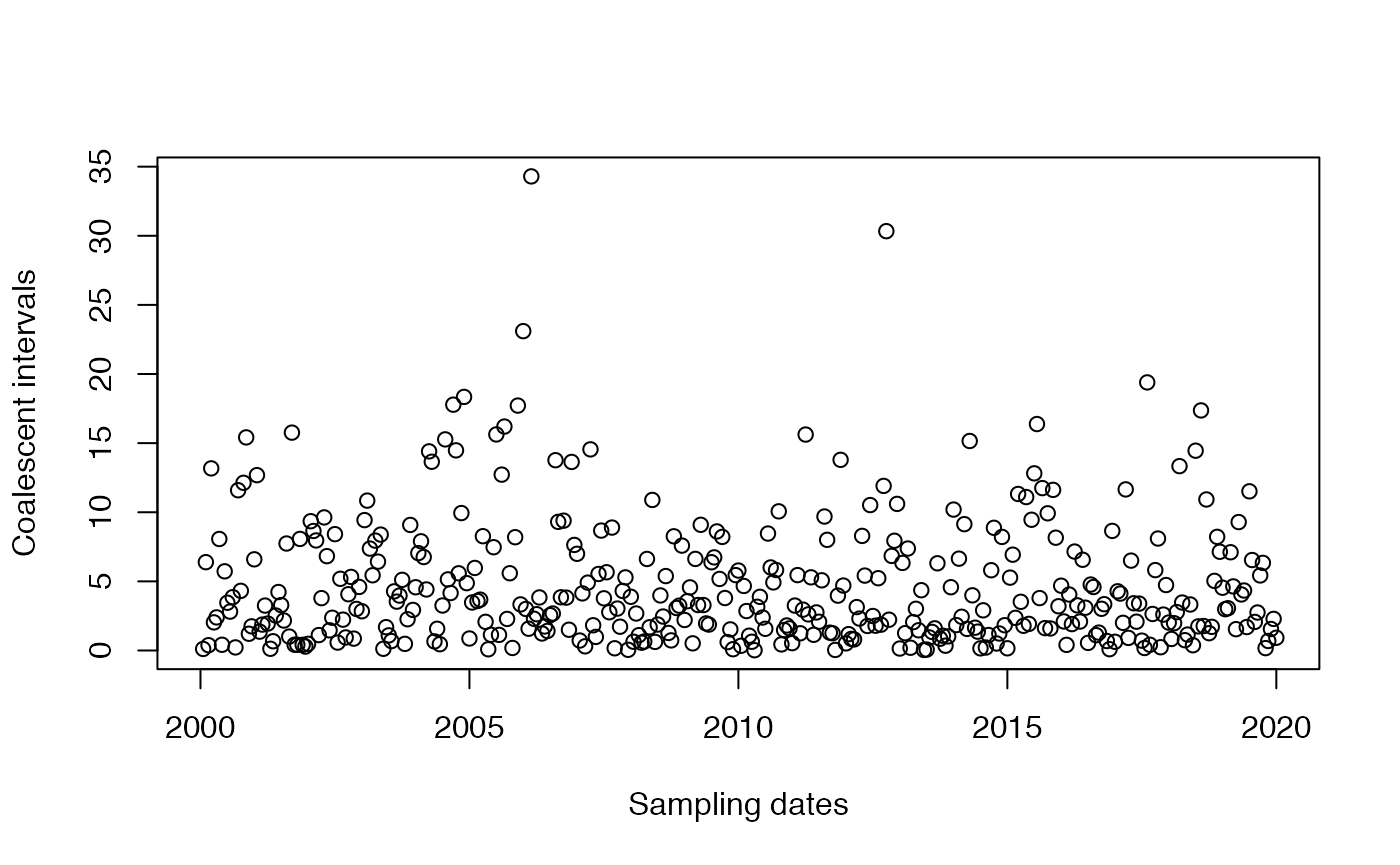
Detection of imports
Imports are likely to have greater than expected if transmission happened only locally. In the case where the location population size is constant equal to , is exponentially distributed with mean . So we can detect imports by looking for coalescent intervals that are greater than this. Since is unknown, we need to estimate it from the data. A simple estimator would be to take the mean of the , but since we suspect that some are affected by imports, it is preferable to use an estimator based on the median: Of course here there are no imports:
res=detectImportsFAST(tree,constant=T)
plot(res,'scatter')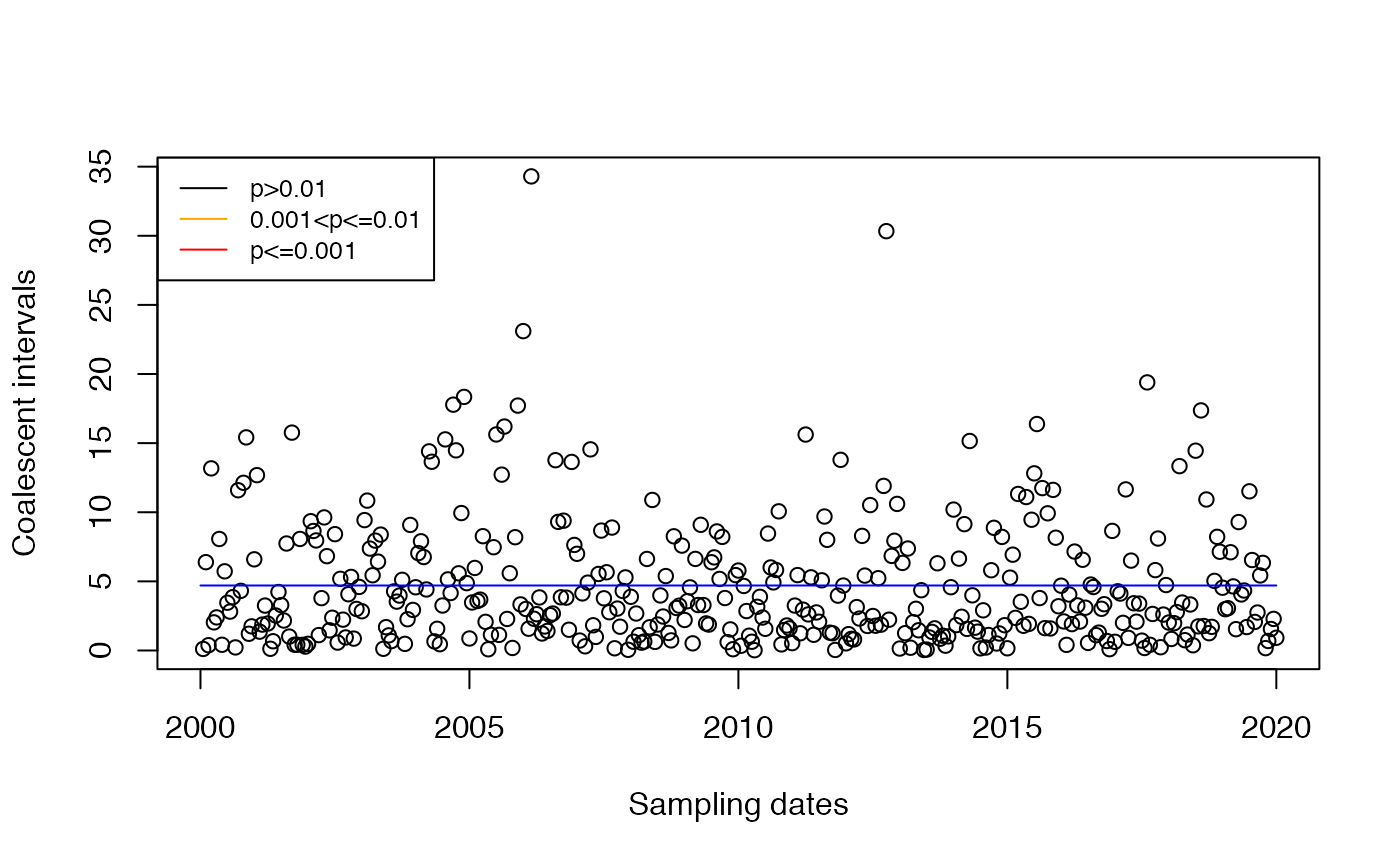
Simulation with varying population size
Now we consider that the effective population size is not constant but is a continuous function of time.
set.seed(0)
Ne=function(t){if (t>2010|t<2005) return(2) else return(10)}
tree=simCoal(seq(2000,2020,0.05),Ne)
plotBoth(tree,Ne)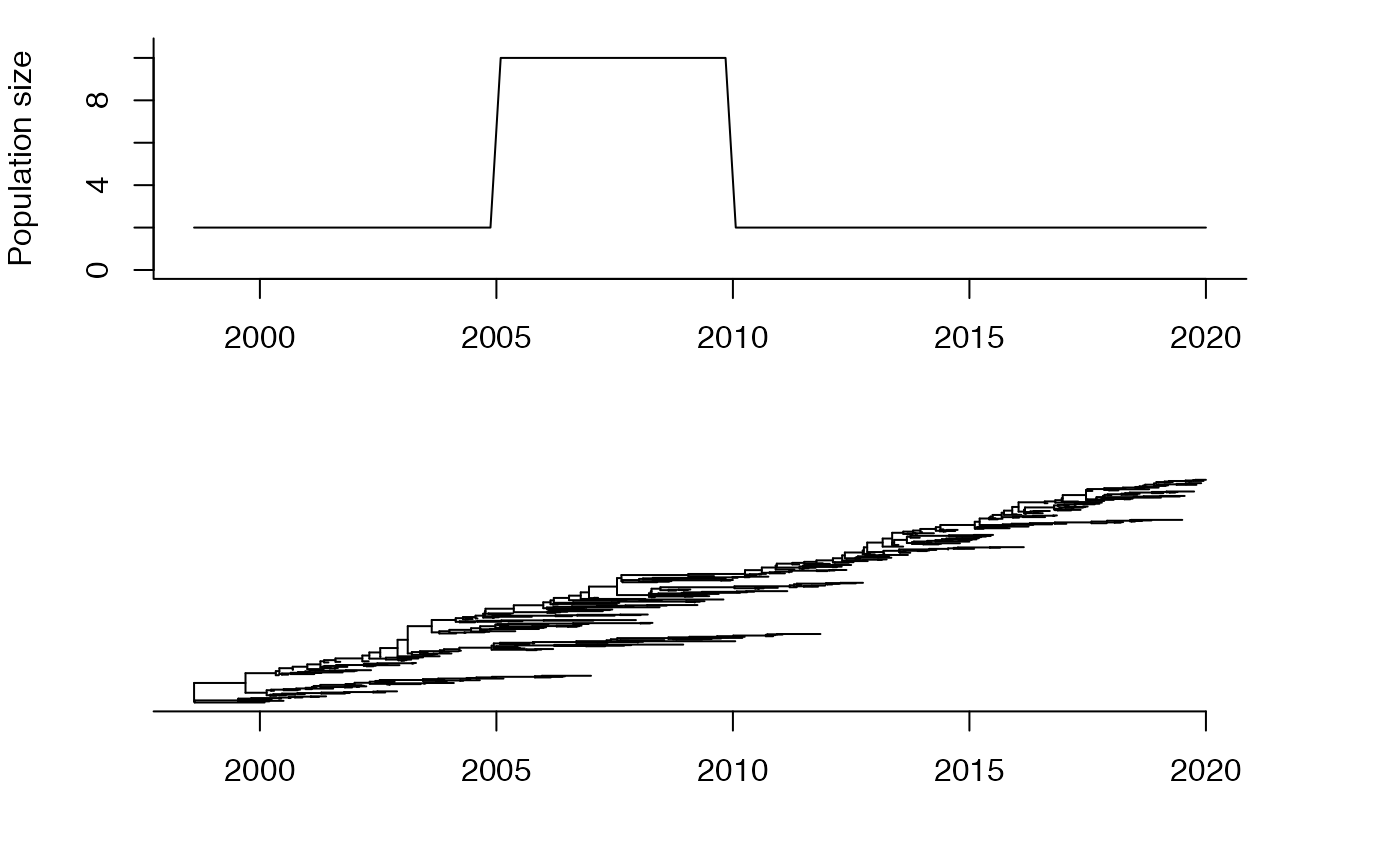
Coalescent intervals
The coalescent intervals are no longer exponential and no longer iid as in the case of a constant population size. But we can still compute them and plot them as before:
plotCoalInt(tree)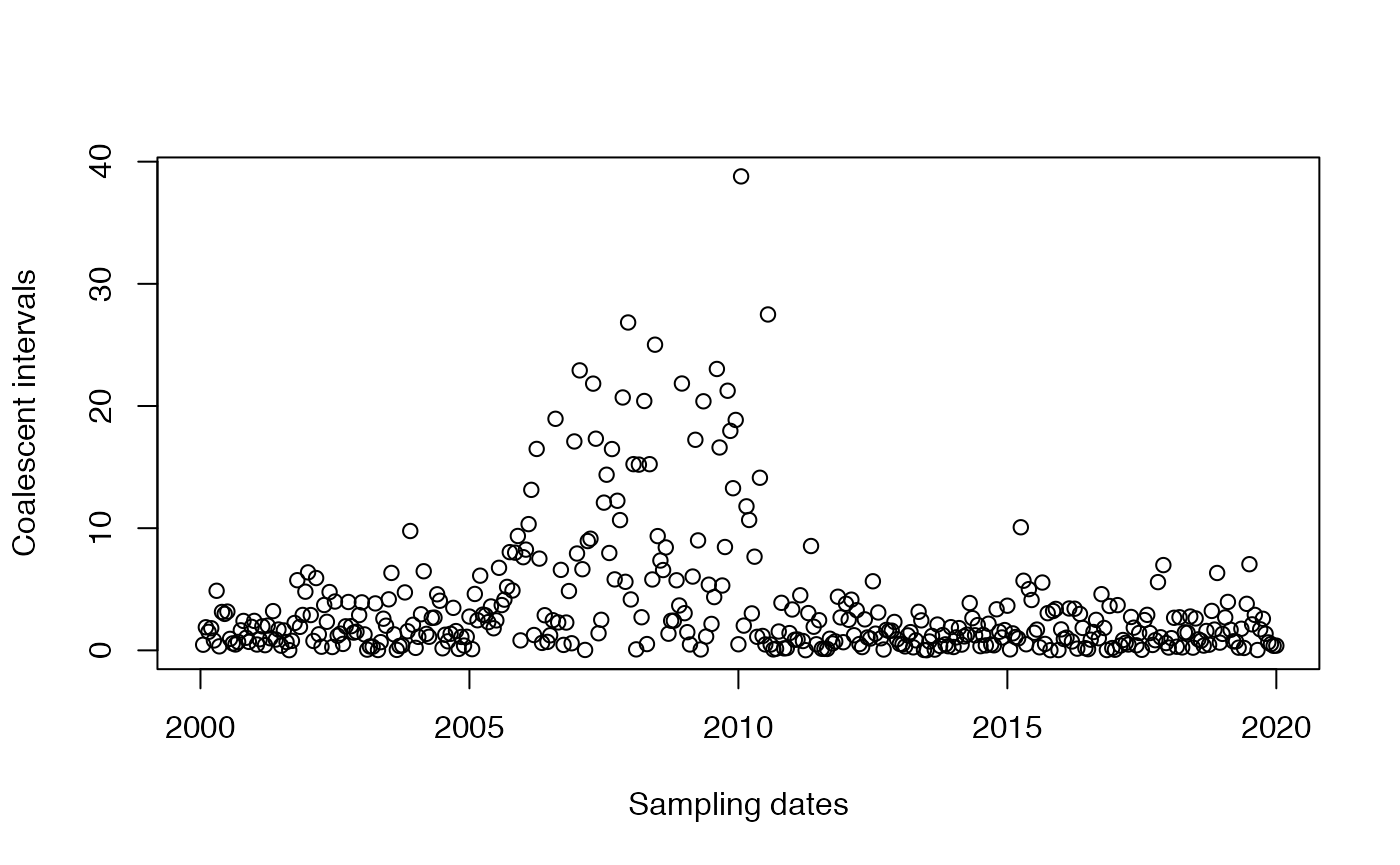
Bad test
If we apply the same test as before, we will get some false positives in the detection of imports:
res=detectImportsFAST(tree,constant=T)
plot(res,'scatter')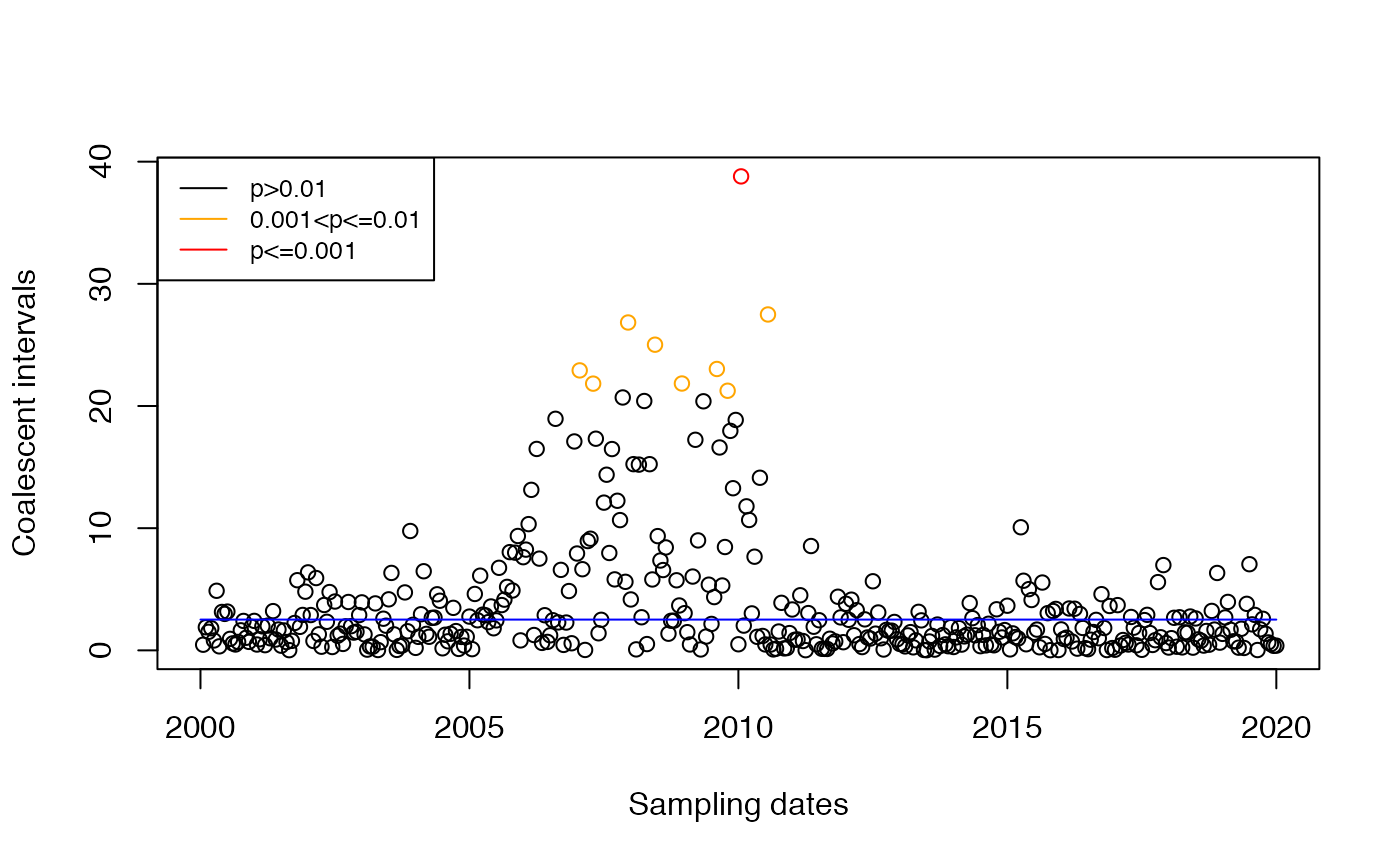
Detection of imports
Outliers in the distribution of against are likely imports. The distribution is unknown and complex in the general case. We use a test in which the distribution is assumed exponential with time-changing mean estimated. This is not quite correct since the distribution is not exponential, but matching the mean is a good first step. As a first attempt, let’s consider the estimator based on the median of other coalescent intervals around the same time:
This estimator is used in the fast test of DetectImports:
res=detectImportsFAST(tree,constant=F,epsilon=1)
plot(res,'scatter')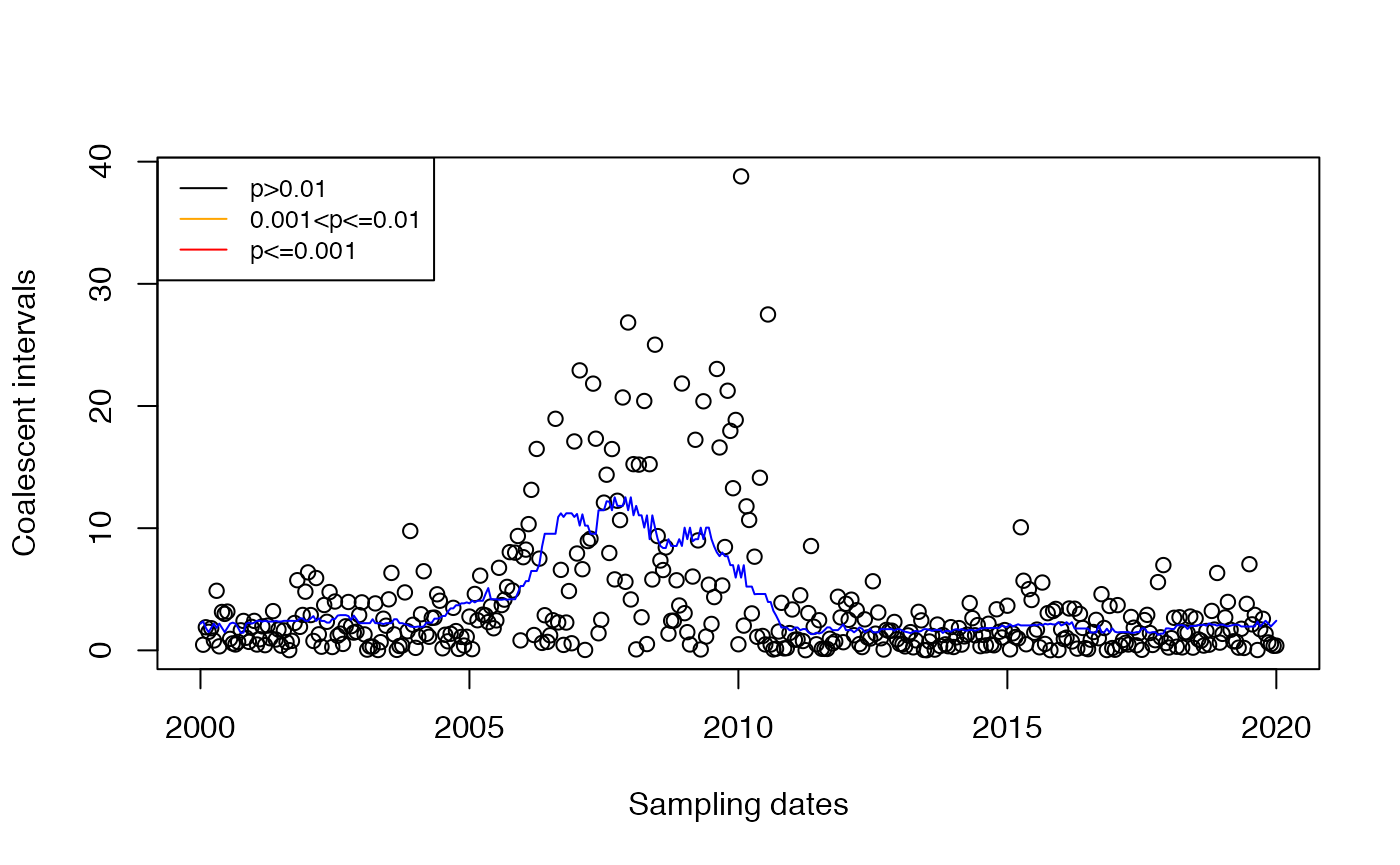
We can see that the false positives are no longer present. In the DetectImports paper, we develop a test based on the same principle but using Bayesian statistics.